-Search query
-Search result
Showing all 37 items for (author: remus & d)

EMDB-15775: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc
Method: single particle / : Davies JS, North RA, Dobson RCJ

PDB-8b01: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc
Method: single particle / : Davies JS, North RA, Dobson RCJ

EMDB-13968: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol
Method: single particle / : North RA, Davies JS, Morado D, Dobson RCJ

PDB-7qha: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol
Method: single particle / : North RA, Davies JS, Morado D, Dobson RCJ

EMDB-27663: 
Open state of RFC:PCNA bound to a 3' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

EMDB-27666: 
Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

EMDB-27667: 
Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

EMDB-27672: 
Closed state of RFC:PCNA bound to a nicked dsDNA
Method: single particle / : Schrecker M, Hite RK

PDB-8dqx: 
Open state of RFC:PCNA bound to a 3' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

PDB-8dqz: 
Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

PDB-8dr0: 
Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

PDB-8dr6: 
Closed state of RFC:PCNA bound to a nicked dsDNA
Method: single particle / : Schrecker M, Hite RK

EMDB-27662: 
Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

EMDB-27668: 
Consensus closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2)
Method: single particle / : Schrecker M, Hite RK

EMDB-27669: 
Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD
Method: single particle / : Schrecker M, Hite RK

EMDB-27670: 
Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD
Method: single particle / : Schrecker M, Hite RK

EMDB-27671: 
Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD
Method: single particle / : Schrecker M, Hite RK

EMDB-27673: 
Open state of RFC:PCNA bound to a nicked dsDNA
Method: single particle / : Schrecker M, Hite RK

PDB-8dqw: 
Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction
Method: single particle / : Schrecker M, Hite RK

PDB-8dr1: 
Consensus closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2)
Method: single particle / : Schrecker M, Hite RK

PDB-8dr3: 
Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD
Method: single particle / : Schrecker M, Hite RK

PDB-8dr4: 
Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD
Method: single particle / : Schrecker M, Hite RK

PDB-8dr5: 
Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD
Method: single particle / : Schrecker M, Hite RK

PDB-8dr7: 
Open state of RFC:PCNA bound to a nicked dsDNA
Method: single particle / : Schrecker M, Hite RK

EMDB-25426: 
Rad24-RFC ADP state
Method: single particle / : Castaneda JC, Schrecker M, Remus D, Hite RK

PDB-7ste: 
Rad24-RFC ADP state
Method: single particle / : Castaneda JC, Schrecker M, Remus D, Hite RK

EMDB-25422: 
Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction
Method: single particle / : Castaneda JC, Schrecker M, Remus D, Hite RK

EMDB-25423: 
Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction
Method: single particle / : Castaneda JC, Schrecker M, Remus D, Hite RK

EMDB-25424: 
Mec3 focus refinement of Rad24-RFC:9-1-1:DNA in the open state
Method: single particle / : Castaneda JC, Schrecker M, Hite RK, Remus D

EMDB-25425: 
Rad24-Rfc4 focus refinement of Rad24-RFC-ADP
Method: single particle / : Castaneda JC, Schrecker M, Hite RK, Remus D

PDB-7st9: 
Open state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction
Method: single particle / : Castaneda JC, Schrecker M, Remus D, Hite RK

PDB-7stb: 
Closed state of Rad24-RFC:9-1-1 bound to a 5' ss/dsDNA junction
Method: single particle / : Castaneda JC, Schrecker M, Remus D, Hite RK

EMDB-23660: 
Cryo-electron tomogram of JCVI-Syn3A
Method: electron tomography / : Lam V, Villa E

EMDB-23661: 
Cryo-electron tomogram of a JCVI_Syn3A cell
Method: electron tomography / : Lam V, Villa E
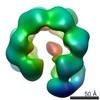
EMDB-3679: 
Full-length complex of S.cerevisiae Cdt1-MCM in apo state.
Method: single particle / : He J, Frigola J, Kinkelin K, Pye VE, Renault L, Douglas M, Remus D, Cherepanov P, Costa A, Diffley JFX
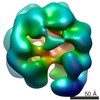
EMDB-3680: 
Full-length complex of S.cerevisiae Cdt1-MCM in ATPgS-bound state.
Method: single particle / : He J, Frigola J, Kinkelin K, Pye VE, Renault L, Douglas M, Remus D, Cherepanov P, Costa A, Diffley JFX
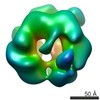
EMDB-3681: 
Full-length complex of S.cerevisiae MCM in ATPgS-bound state.
Method: single particle / : He J, Frigola J, Kinkelin K, Pye VE, Renault L, Douglas M, Remus D, Cherepanov P, Costa A, Diffley JFX
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
